What are the most common pests and diseases when growing plants in controlled environments?
Growing plants in the lab, whether this is in growth chambers or in greenhouses, isn’t always easy. It is important to look for pests and diseases regularly to make sure the plants are healthy and to take care of any problem as soon as possible to avoid its spreading.🌱
1️⃣Fungus gnats

Fungus gnats are mosquito-like insects from the Sciaridae family. These are probably the most common pests when growing plants in controlled environments. Usually, they are easy to spot, as the adults are often found flying around. Even though the adults are typically harmless they can, in some cases, carry diseases among plants. The real problem when it comes this pest is their larvae, which are known to eat the plant’s roots and leaves, posing a heavy threat to seedlings rather than bigger plants. Plants infested with fungus gnats display symptoms such as eaten leaves and very small root systems. Fungus gnats can be controlled using several methods, such as sticky traps, insecticides or biological control (nematodes, carnivorous plants, mites, etc).
2️⃣Thrips

Thrips are tiny, slender insects with fringed wings which belong to the order Thysanoptera. They are easy to spot as the plants begin to show white and discolored spots in their leaves. This happens because thrips feed by puncturing the epidermal layer of the plant’s leaf, sucking out the cell contents. Thrips can be vectors for plant diseases and can be controlled using insecticides or biological control.
3️⃣Aphids

Aphids are sap-sucking insects which belong to the superfamily Aphidoidea. Even though they do not commonly appear in growth chambers, aphids are a common pest in greenhouses. Plants infested with aphids usually display symptoms such as decreased growth rates, yellowing, browning, stunted growth, wilting, and low yields. Aphids are also known to frequently transmit plant viruses to their hosts and to excrete honeydew which often leads to the growth of fungi - sooty moulds - in the host plant. It can be difficult to control an aphid infestation due to its ability to rapidly increase in numbers by asexual reproduction. The most common ways of controlling this pest is by using insecticides and biological controls.
4️⃣Powdery mildew

Powdery mildew is a fungal disease caused by many different species of ascomycete fungi of the order Erysiphales. Powdery mildew is very easy to identify as infected plants display white powdery spots on its leaves and stems. As fungi usually do, powdery mildew grows well in humid environments and produces spores which can be carried to other plants to spread the infection. Usually, this disease is controlled using fungicides.
Text by Clarisse Zigue PhD Student EpiSeedLink Marie Skłodowska-Curie Actions
Frontal image by <a href="https://www.freepik.es/foto-gratis/verduras-organicas-frescas-cultivadas-interiores-hidroponia-generadas-ia_42649480.htm#page=4&query=growing%20plants%20in%20controlled%20environments&position=3&from_view=search&track=ais">Imagen de vecstock</a> en Freepik
ChIP seq
The fellows just met in Madrid on the week of the 18th-22nd of September to learn how to perform a ChIP-seq on Brassica napus. They first went in-depth into the theory and then they went hands-on the protocol.
But… What is ChIP-seq?
ChIP-seq is a tool to investigate the epigenomes of multiple cell types and their underlying mechanisms. It is based on chromatin immunoprecipitation (ChIP) which is a protein-DNA binding in vivo assay technique. The ChIP antibodies are specific to our protein or nucleosome of interest. ChIP is followed by sequencing (the –seq part of ChIP-seq), which allows to identify and sequence our enriched DNA fragments (Park, 2009).
There are four basic steps for ChIP, as you can see in the image below. Firstly, crosslinking is the way to improve the connection between protein and chromatin to preserve the interaction for further antibody targeting. There are two common methods to do crosslinking, using formaldehyde or ultraviolet (UV) light. For this ChIP-seq course, we used 1% formaldehyde. After crosslinking, we need to extract chromatin, and shearing/sonicate them. Shearing step to solubilize the chromatin which is very important to get enough chromatin yield. In addition, a proper shearing strategy to generate desirable DNA size (300bp-500bp) is good for immunoprecipitation and signal detection. The next step is to remove these fragments without the connection between the targeted protein and chromatin. To achieve this, we need to use antibodies to bind with targeted proteins where the antibody can stick to magnetic beads. So, the non-target proteins can be washed out, followed by eluting the targeted protein from the beads. Then, de-crosslinking means removing the proteins from the DNA to make the purified DNA accessible easily. Lastly, indexing barcodes will be added to the purified DNA for individual sample identification. If the quality control test shows positive results, the library could be used for ChIP-seq.
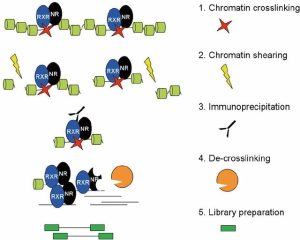
(Daniel et al., 2014)
From this we understand how ChIP sequencing is carried out and the steps involved in successful preparation of a library for sequencing, further using various sequencing platforms sequencing of the prepared libraries can be carried out.
With excellent guidance by Dr. Pedro Crevillén and Dr. Jose A. Abelenda, fellows of the EpiSeedLink doctoral network were skillfully trained in ChIP sequencing. This will immensely help each of the fellows in the next phase of their PhD journey, where they will incorporate the knowledge gained in practical terms.
Each fellow will use ChIP sequencing technique to uniquely benefit their research project, resulting in generating enormous data that will help the research community in understanding the dynamics of both Arabidopsis and Brassica napus genome.
Text by Shreyas Padmanabha Sharma Beedubail, PhD Student EpiSeedLink Marie Skłodowska-Curie Actions
References:
- Daniel, B., Balint, B. L., Nagy, Z. S., & Nagy, L. (2014). Mapping the genomic binding sites of the activated retinoid X receptor in murine bone marrow-derived macrophages using chromatin immunoprecipitation sequencing. Steroid Receptors: Methods and Protocols, 15-24.
- Park, P. J. (2009). ChIP–seq: advantages and challenges of a maturing technology. Nature reviews genetics, 10(10), 669-680.
Frontal image by <a href="https://www.freepik.es/foto-gratis/especialista-biotecnologia-laboratorio-realizando-experimentos_44133702.htm#query=ChIP%20seq%20DNA%20protein&position=0&from_view=search&track=ais">Freepik</a>



