Studying genes and transformation plants
When we want to study the role of a gene, we can alter its expression; for example: silencing or suppressing the gene or improving its expression using a strong and constitutively expressed promoter.

Genetic engineering is a powerful research tool, and it also has the potential to improve the agronomic performance of crops, for these reasons, it has been largely studied and a lot of different techniques have been developed in recent years.
So how do we transform plants?
Transformation is a complex process, it may be transient or stable, depending on the biological question and purpose.
Generally, transformation involves:
- the preparation of explants,
- delivery of genes of interest into plant cells mainly via:
- Agrobacterium or
- biolistic-mediated methods,
- the selection and regeneration of transgenic or gene-edited plants.
Text by Stefania Paltrinieri, PhD Student EpiSeedLink Marie Skłodowska-Curie Actions
Frontal image by <a href="https://www.freepik.es/vector-gratis/concepto-biotecnologia-gradiente-investigador_13176503.htm#fromView=search&page=1&position=8&uuid=dd08f069-784b-4102-a785-4cd12126319a">Imagen de freepik</a>
How do plants tell time?
Continuing thinking about plant curiosities, another question is: How do plants tell time?
Some flowers open their petals at specific times of the day. Some plants move their leaves up during the day, and move their leaves down during the night. These and other plants' events happen at specific times of the day and follow the internal clock of the plants.
The internal clock of plants is called the circadian clock. This is a molecular mechanism independent of environmental cues. Then, even if the plant is in constant light or darkness, the plant will still move its leaves up during the period of the day, and down during the period of the night, for example.
The main function of the circadian clock in plants is to anticipate the day and night and the cycles of temperature during one day. So everything happens at the right time. When the plants anticipate wrong… well, they may have a decrease in productivity, survival, and fitness.
The main references are:
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6466001/
https://plantsinmotion.bio.indiana.edu/plantmotion/movements/leafmovements/bean/beansleep.html
Text and video by Maira Marins Dourado, PhD Student EpiSeedLink Marie Skłodowska-Curie Actions
Frontal image by <a href="https://www.freepik.es/vector-gratis/ilustracion-concepto-resorte-adelante_137950301.htm#fromView=search&page=3&position=26&uuid=dbed3ed9-1c24-4a59-85d1-919eeef55494">Imagen de storyset en Freepik</a>
Do the plants sleep?
Thinking about plant curiosities? One question is: Do the plants sleep?
In essence, sleep is defined as a behavior. We sleep when we are in a state where a weak disturbance will not awake us, but a strong disturbance will revert this state rapidly.
However, we can explain sleep in animals in other ways. The period of sleep is to save energy, repair the body, and direct the energy to functions less active when we are awake.
During the evening, plants do very different activities compared to the day.
When it is day, plants focus on absorbing light and producing sugar, for example.
When it is night, plants focus on consuming sugar, producing proteins, and
growing.
3- As different as day and night
Then... do plants sleep? Sleep is mainly described as animal behaviour, that appeared very early in animal evolution.
However, different uses of energy happen to plants during the day and night and when animals are asleep and awake.
So, plants don’t sleep like us. But, like humans, do different activities during the evening compared to the day.
The main references are:
https://www.nature.com/articles/s41583-018-0098-9
https://onlinelibrary.wiley.com/doi/10.1111/j.1365-3040.2007.01708.x
Text and video by Maira Marins Dourado, PhD Student EpiSeedLink Marie Skłodowska-Curie Actions
Frontal image by <a href="https://www.freepik.es/foto-gratis/mujer-descansando-hierba_2042318.htm#page=9&query=plantas%20durmiendo&position=31&from_view=search&track=ais&uuid=eb8662d1-a889-4e00-b157-94e6f57cae34">Freepik</a>
Immunoflorescence technique used in epigenetic research
As researchers, most of us currently perform Western Blots and PCR, but working in the field of epigenetics, we can use other techniques.
Immunofluorescence is an immunohistochemistry technique that uses fluorophores to visualize cellular antigens, such as proteins.
This technique can be utilized to visualize the localization of various cellular components within cells and within cell compartments.
In epigenetics, it's largely used to observe the distribution of different histone modifications or histone variants and so to have an idea of the nuclear organization and chromatin landscape.
For instance, this allows us to see heterochromatic and euchromatic regions within the nucleus and how their distribution changes in different conditions.
The key steps of the protocol are:
-
- Fixation with formaldehyde, to prevent the degradation of your sample
- Incubation with a primary antibody that specifically recognizes the protein of interest or the PTM of interest
- Incubation with a secondary antibody coupled to a fluorophore and, if your protein interacts with DNA don't forget to stain your sample with DAPI!
- Observation with a fluorescence microscope or a confocal microscope
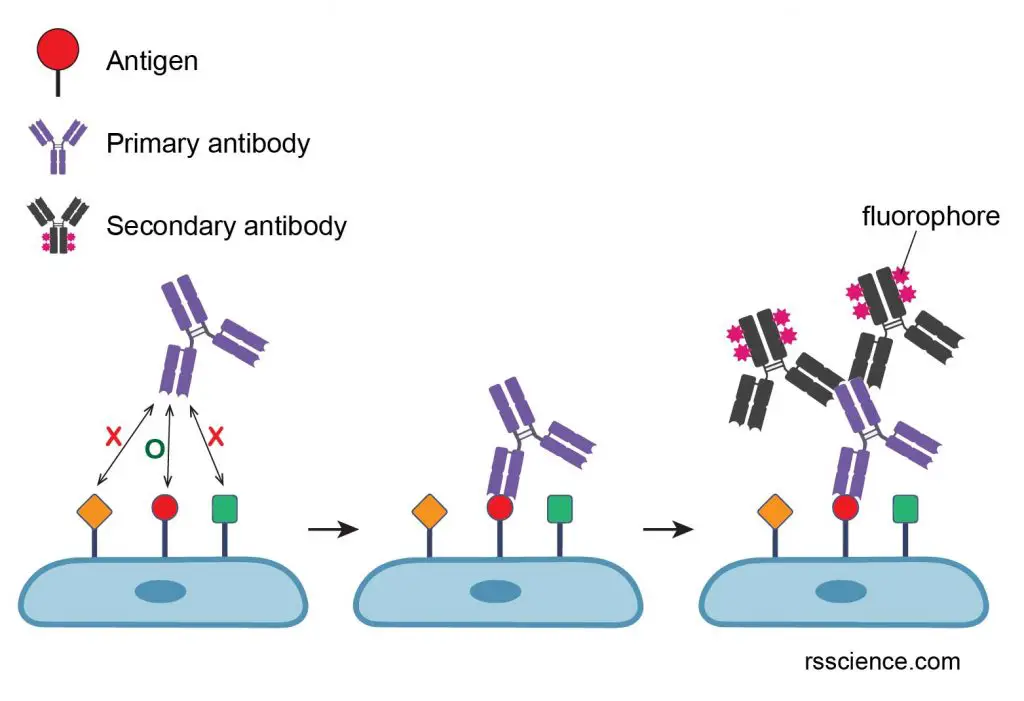
Text by Stefania Paltrinieri, PhD Student EpiSeedLink Marie Skłodowska-Curie Actions
Frontal image by Nicola Ferrari
Evolution of Brassica napus
- Brassica napus is a crop species of economic importance. It is cultivated worldwide as oil and livestock crops.
- It belongs to the genus Brassica, tribe Brassiceae of the family Brassicaceae.
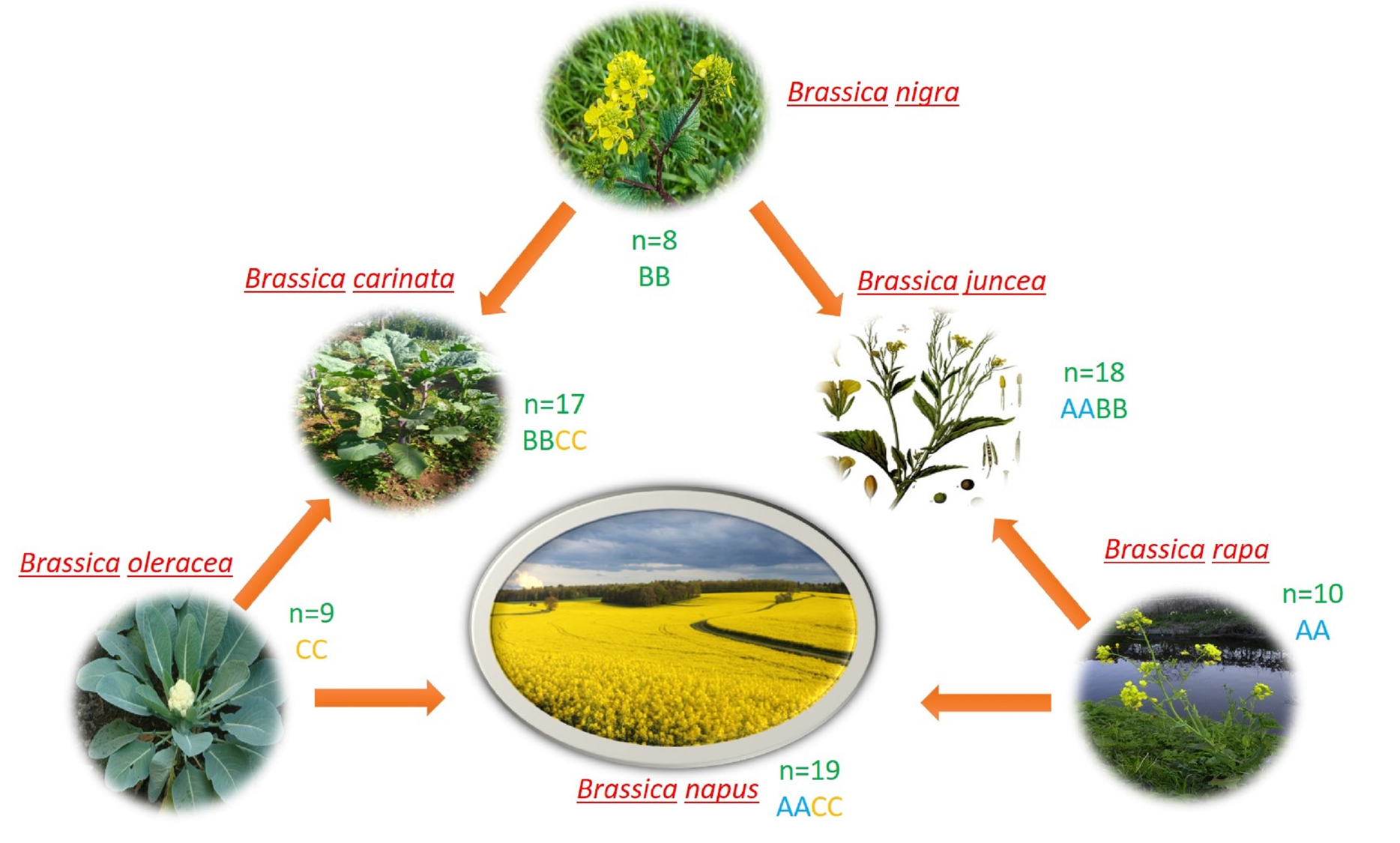
- The genus Brassica includes many important crops. Among them, relationship of six species formed the model of U’s triangle, with three basic diploid species B. rapa (A genome, n = 10), B. oleracea (C genome, n = 9) and B. nigra (B genome, n = 8) gave rise to three amphidiploid species B. napus (AC genome, n = 19), B. juncea (AB genome, n = 18) and B. carinata (BC genome, n = 17).
Text by Shreyas Padmanabha Sharma Beedubail, PhD Student EpiSeedLink Marie Skłodowska-Curie Actions
Source:
- X. Wang and C. Kole (eds.), The Brassica rapa Genome, Compendium of Plant Genomes, 2015, DOI 10.1007/978-3-662-47901-8_1
- By Kumar83 - Own work, CC0, https://commons.wikimedia.org/w/index.php?curid=8946874
- By Ztmcfaul - Self-photographed, Public Domain, https://commons.wikimedia.org/w/index.php?curid=84890196
- https://encrypted-tbn0.gstatic.com/licensed-image?q=tbn:ANd9GcTEkOpTlpXlCIqz0w1asz-Y9NItAuLu5wliax0MYZqstG8qNP2YPdLseX5j6S7Q16ulNpBkr6GLz34sGQ0
- By Walther Otto Müller - List of Koehler Images, Public Domain, https://commons.wikimedia.org/w/index.php?curid=255508
- By TeunSpaans - Own work, CC BY-SA 3.0, https://commons.wikimedia.org/w/index.php?curid=96733
- By Photo: Myrabella / Wikimedia Commons, CC BY-SA 4.0, https://commons.wikimedia.org/w/index.php?curid=33781153
- https://en.wikipedia.org/wiki/Triangle_of_U
Chromatin Players: SWI/SNF Chromatin Remodeling Complex
Chromatin, the intricate combination of DNA and histone proteins, plays a pivotal role in governing nuclear processes. In this article, our focus is on the dynamic SWI/SNF chromatin remodeling complex. Essential to the epigenetic regulation of numerous nuclear processes, ranging from transcription to DNA repair and replication, SWI/SNF stands as a key player in the complex world of chromatin dynamics.
Chromatin Structure and Dynamics: A Brief Overview
Nucleosomes, the basic units of chromatin, create a barrier to protein access within the genome. However, these nucleosomes are not static; their occupancy and positioning are subject to alteration by chromatin remodeling complexes (CRCs). Using energy derived from ATP hydrolysis, CRCs, including the SWI/SNF complex, disrupt electrostatic interactions between histones and DNA.
SWI/SNF: A Unique Chromatin Remodeling Player
Among the four main classes of CRCs found in yeast, animals, and plants (Imitation Switch, SWI; Inositol-Requiring 80, INO80; Chromodomain Helicase DNA-Binding, CHD; and Mating Type Switch Deficient Sucrose Nonfermenting, SWI/SNF), the SWI/SNF complex stands out. Its distinct ability to modify the accessibility of genomic DNA within chromatin sets it apart. SWI/SNF achieves this by not only sliding the histone octamer along the DNA but also by ejecting it, influencing crucial nuclear processes like transcription, DNA repair, and replication.
SWI/SNF Architecture: Modules and Functions
SWI/SNF CRCs are colossal protein assemblies, weighing between 1 and 2 megadaltons. Comprising multiple conserved and interconnected subunits, including a central ATPase, these complexes are organized into three modules: the motor module, the actin-related protein (ARP) module, and the base module.
- Motor Module: This powerhouse hydrolyzes ATP and disrupts DNA-histone contacts, playing a pivotal role in reshaping chromatin structure.
- ARP Module: Serving as a connector between the motor and base modules, ARP enhances the coupling of ATP hydrolysis to DNA translocation, ensuring efficient chromatin remodeling.
- Base Module: Engaging with histones, this module's interactions are influenced by histone posttranslational modifications and specific DNA sequence features, adding another layer of complexity to the chromatin remodeling process.
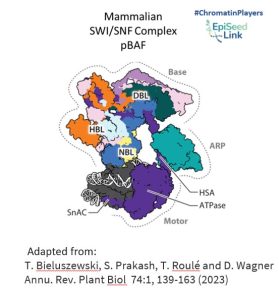
Activities of SWI/SNF Complexes in the Nucleus
Within the nucleus, SWI/SNF remodelers orchestrate transcriptional regulation at various levels. They participate in modulating enhancer activity, recruiting the general transcriptional machinery, and facilitating the transition from paused to elongating RNA polymerase.
SWI/SNF is recruited to genomic DNA by transcription factors (TFs), unable to bind DNA directly. Pioneering TFs uniquely access DNA wrapped in nucleosomes, recruiting SWI/SNF to reduce nucleosome occupancy or move nucleosomes away from enhancers. This process enables the binding of additional TFs required for complete transcriptional output.
Additionally, SWI/SNF facilitates the access of RNA polymerase II to the promoter, contributing to the initiation of transcription. Notably, SWI/SNF complexes push the +1 nucleosome away from the transcription start site (TSS), enabling the recruitment of RNA polymerase II and the general transcriptional machinery.
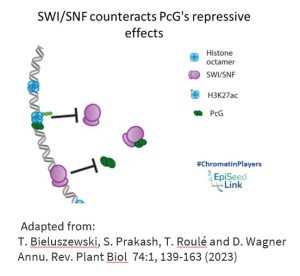
SWI/SNF in Epigenetic Regulation: Interplay with Polycomb Group Proteins
Polycomb group (PcG) epigenetic regulators, known for silencing transcriptional programs, intersect with SWI/SNF in a fascinating manner. PcG complexes PRC2 and PRC1 deposit repressive histone modifications, including H3K27me3. SWI/SNF CRCs, acting as molecular gatekeepers, eject PcG complexes and impede their access to chromatin at numerous loci. This dynamic interplay between PcG and SWI/SNF contributes to shaping the accessible chromatin landscape during developmental processes in both animals and plants.
Unraveling SWI/SNF Complexity in Plants: Insights from Arabidopsis
In the plant kingdom, understanding the classification and composition of SWI/SNF complexes has been a challenge. In Arabidopsis thaliana three types of SWI/SNF ATPase has been identified: BRAHMA (BRM/CHR2), SPLAYED (SYD/CHR3), MINUSCULE1 (MINU1/CHR12), and its closed related MINUSCULE2 (MINU2/CHR23). These ATPases, though conserved in the ATPase domain, exhibit diverse domain structures at the N-terminal and C-terminal regions, suggesting functional divergence. They also form different complexes (BAS, SAS or MAS) by interacting with different groups of common or specific subunits.
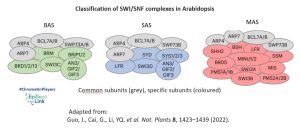
Mutant analysis of BRM, SYD, MINU1, and MINU2 offers a glimpse into the crucial role these ATPases play in plant development. Single mutants, such as brm and syd, exhibit pleiotropic defects—reduced plant size, slow growth, curled rosette leaves, aberrant flower development, and sterility. BRM and SYD occupy thousands of common target genes and exhibit similar binding patterns, indicating that the two enzymes are functionally related.
The brm syd double mutant faces embryonic lethality, emphasizing the essential nature of these ATPases in early embryo development.
Loss-of-function minu1 and minu2 single mutants are morphologically indistinguishable from the wild type, while the minu1/2 (minu1 and minu2) double mutant exhibits embryonic lethal phenotypes. The weak minu1/2 double mutant forms small and bushy plants that have defects in maintenance of both root and shoot apical meristems. These results suggest that the Arabidopsis SWI/SNF ATPases have specific functions in regulating development, although the mechanisms underlying the specificity are mainly unknown.
Analyzing the phenotype of the mutants of three types of SWI/SNF complex concluded that three classes of SWI/ SNF complexes have both overlapping and specific functions in the regulation of various developmental processes, including the development of leaves, gametophytes, embryos and flowers.
Moreover, it has been shown that the three types of SWI/SNF complexes differently regulate chromatin accessibility, providing a plausible explanation for the specific functions of different SWI/SNF complexes.
Analysis of mutants representing three types of SWI/SNF complexes revealed overlapping and specific functions in the regulation of diverse developmental processes, encompassing leaf, gametophyte, embryo, and flower development. Additionally, distinct regulatory roles were observed as the three SWI/SNF complex types differentially influenced chromatin accessibility, providing insight into their specific functionalities.
Examining genomic distribution patterns, all three plant SWI/SNF complexes predominantly bind to the transcription start site (TSS)-flanking regions of protein-coding genes, indicating shared intrinsic features facilitating this binding. However, the BAS complex primarily targets the TSS-flanking region, while SAS and MAS extend their distribution to the upstream distal and downstream intragenic regions, respectively. This divergence is suspected to arise from varying binding affinities for active histone modifications H3ac and H3K4me3, which are enriched at the intragenic region.
The distinct binding abilities of BAS, SAS, and MAS for active histone modifications likely dictate their selective binding to different gene regions. Consequently, active histone modifications regulate the function of these complexes at both the chromatin-binding and nucleosome remodeling levels, although the precise molecular mechanisms remain elusive.
The proposed mechanism involves specific modified-histone readers within SWI/SNF complexes, responsible for recognizing histone modifications and determining the diverse regulation of nucleosome remodeling activities across the three SWI/SNF complex classes. This interplay between complex composition and histone modification patterns highlights the evolution of SWI/SNF complexes with a flexible selection of distinct histone readers, showcasing their fine-tuned adaptation to transcriptional regulation in Arabidopsis.
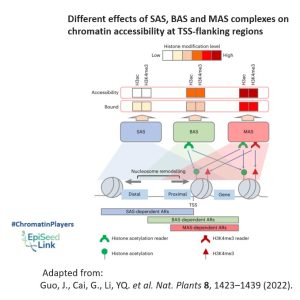
In conclusion, this review highlights the vital role of the SWI/SNF chromatin remodeling complex in Arabidopsis development, shedding light on the distinct functions of its ATPase subunits. As we explore chromatin accessibility and genomic distribution patterns, it becomes evident that further research is essential to uncover the intricate mechanisms governing SWI/SNF-mediated transcriptional regulation in plants.
References
- Bieluszewski, T., Prakash, S., Roulé, T., Doris Wagner, D. The Role and Activity of SWI/SNF Chromatin Remodelers. Annu. Rev. Plant Biol 74:1, 139-163 (2023) https://doi.org/10.1146/annurev-arplant-102820-093218
- Guo, J., Cai, G., Li, YQ. et al. Comprehensive characterization of three classes of Arabidopsis SWI/SNF chromatin remodeling complexes. Nat. Plants 8, 1423–1439 (2022). https://doi.org/10.1038/s41477-022-01282-z
Text by Carlos Gámez Álvarez, PhD Student EpiSeedLink Marie Skłodowska-Curie Actions
Frontal image from https://bit.ly/3RrxEP7
What are the most common pests and diseases affecting Brassica Napus in the field?

✅Blackleg
The most common fungal disease affecting oilseed rape (B. napus) in European fields is Leptosphaeria maculans, causing blackleg. This disease manifests as dark elongated lesions on stems and leaves, leading to lodging and reduced yields. It's a significant concern in Europe's oilseed rape cultivation, impacting plants at various growth stages. The fungus survives in crop residues, infecting new crops through spores produced on infected debris. Effective management strategies include planting resistant cultivars, crop rotation, and integrated pest management, which may involve fungicide application based on local recommendations. The disease's severity varies across regions and years, underscoring the importance of staying informed and adopting a range of practices to mitigate its impact on oilseed rape crops in Europe.

✅Sclerotinia Stem Rot
Another common disease affecting oilseed rape in European fields is Sclerotinia Stem Rot, caused by the fungus Sclerotinia sclerotiorum, commonly known as "white mold." This disease presents as white cottony mycelial growth on stems, leaves, and pods, leading to wilting, lodging, and premature plant death. Sclerotinia Stem Rot is particularly concerning in regions with cool and humid conditions. The pathogen overwinters as hard resting structures called sclerotia and spreads through wind and rain-dispersed spores. Effective management involves using resistant cultivars, implementing crop rotation, improving air circulation through wider row spacing, and applying fungicides preventively during periods of high humidity. Given its impact on oilseed rape cultivation, especially in Europe's favorable conditions, integrated disease management strategies are vital for reducing the disease's significant economic and yield-related consequences.

✅Cabbage Stem Flea Beetle (Psylliodes chrysocephalus)
The most prevalent insect pest affecting B. napus (oilseed rape) in European fields is the Cabbage Stem Flea Beetle (Psylliodes chrysocephalus). These beetles cause damage by feeding on seedlings, resulting in "shot hole" damage, and larvae can harm stems, potentially causing lodging. Active during spring, they overwinter as adults and lay eggs in the soil. Effective management involves early planting, cultural practices like trap crops, and judicious insecticide use when necessary. While the Cabbage Stem Flea Beetle is a major concern, the impact of insect pests on oilseed rape varies by region and climate, emphasizing the importance of vigilance and integrated pest management for maintaining healthy crops.

Text by Kingsley Onyinye Ibeabuchi, PhD Student EpiSeedLink Marie Skłodowska-Curie Actions
What are the most common pests and diseases when growing plants in controlled environments?
Growing plants in the lab, whether this is in growth chambers or in greenhouses, isn’t always easy. It is important to look for pests and diseases regularly to make sure the plants are healthy and to take care of any problem as soon as possible to avoid its spreading.🌱
1️⃣Fungus gnats

Fungus gnats are mosquito-like insects from the Sciaridae family. These are probably the most common pests when growing plants in controlled environments. Usually, they are easy to spot, as the adults are often found flying around. Even though the adults are typically harmless they can, in some cases, carry diseases among plants. The real problem when it comes this pest is their larvae, which are known to eat the plant’s roots and leaves, posing a heavy threat to seedlings rather than bigger plants. Plants infested with fungus gnats display symptoms such as eaten leaves and very small root systems. Fungus gnats can be controlled using several methods, such as sticky traps, insecticides or biological control (nematodes, carnivorous plants, mites, etc).
2️⃣Thrips

Thrips are tiny, slender insects with fringed wings which belong to the order Thysanoptera. They are easy to spot as the plants begin to show white and discolored spots in their leaves. This happens because thrips feed by puncturing the epidermal layer of the plant’s leaf, sucking out the cell contents. Thrips can be vectors for plant diseases and can be controlled using insecticides or biological control.
3️⃣Aphids

Aphids are sap-sucking insects which belong to the superfamily Aphidoidea. Even though they do not commonly appear in growth chambers, aphids are a common pest in greenhouses. Plants infested with aphids usually display symptoms such as decreased growth rates, yellowing, browning, stunted growth, wilting, and low yields. Aphids are also known to frequently transmit plant viruses to their hosts and to excrete honeydew which often leads to the growth of fungi - sooty moulds - in the host plant. It can be difficult to control an aphid infestation due to its ability to rapidly increase in numbers by asexual reproduction. The most common ways of controlling this pest is by using insecticides and biological controls.
4️⃣Powdery mildew

Powdery mildew is a fungal disease caused by many different species of ascomycete fungi of the order Erysiphales. Powdery mildew is very easy to identify as infected plants display white powdery spots on its leaves and stems. As fungi usually do, powdery mildew grows well in humid environments and produces spores which can be carried to other plants to spread the infection. Usually, this disease is controlled using fungicides.
Text by Clarisse Zigue PhD Student EpiSeedLink Marie Skłodowska-Curie Actions
Frontal image by <a href="https://www.freepik.es/foto-gratis/verduras-organicas-frescas-cultivadas-interiores-hidroponia-generadas-ia_42649480.htm#page=4&query=growing%20plants%20in%20controlled%20environments&position=3&from_view=search&track=ais">Imagen de vecstock</a> en Freepik
ChIP seq
The fellows just met in Madrid on the week of the 18th-22nd of September to learn how to perform a ChIP-seq on Brassica napus. They first went in-depth into the theory and then they went hands-on the protocol.
But… What is ChIP-seq?
ChIP-seq is a tool to investigate the epigenomes of multiple cell types and their underlying mechanisms. It is based on chromatin immunoprecipitation (ChIP) which is a protein-DNA binding in vivo assay technique. The ChIP antibodies are specific to our protein or nucleosome of interest. ChIP is followed by sequencing (the –seq part of ChIP-seq), which allows to identify and sequence our enriched DNA fragments (Park, 2009).
There are four basic steps for ChIP, as you can see in the image below. Firstly, crosslinking is the way to improve the connection between protein and chromatin to preserve the interaction for further antibody targeting. There are two common methods to do crosslinking, using formaldehyde or ultraviolet (UV) light. For this ChIP-seq course, we used 1% formaldehyde. After crosslinking, we need to extract chromatin, and shearing/sonicate them. Shearing step to solubilize the chromatin which is very important to get enough chromatin yield. In addition, a proper shearing strategy to generate desirable DNA size (300bp-500bp) is good for immunoprecipitation and signal detection. The next step is to remove these fragments without the connection between the targeted protein and chromatin. To achieve this, we need to use antibodies to bind with targeted proteins where the antibody can stick to magnetic beads. So, the non-target proteins can be washed out, followed by eluting the targeted protein from the beads. Then, de-crosslinking means removing the proteins from the DNA to make the purified DNA accessible easily. Lastly, indexing barcodes will be added to the purified DNA for individual sample identification. If the quality control test shows positive results, the library could be used for ChIP-seq.
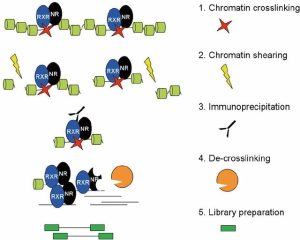
(Daniel et al., 2014)
From this we understand how ChIP sequencing is carried out and the steps involved in successful preparation of a library for sequencing, further using various sequencing platforms sequencing of the prepared libraries can be carried out.
With excellent guidance by Dr. Pedro Crevillén and Dr. Jose A. Abelenda, fellows of the EpiSeedLink doctoral network were skillfully trained in ChIP sequencing. This will immensely help each of the fellows in the next phase of their PhD journey, where they will incorporate the knowledge gained in practical terms.
Each fellow will use ChIP sequencing technique to uniquely benefit their research project, resulting in generating enormous data that will help the research community in understanding the dynamics of both Arabidopsis and Brassica napus genome.
Text by Shreyas Padmanabha Sharma Beedubail, PhD Student EpiSeedLink Marie Skłodowska-Curie Actions
References:
- Daniel, B., Balint, B. L., Nagy, Z. S., & Nagy, L. (2014). Mapping the genomic binding sites of the activated retinoid X receptor in murine bone marrow-derived macrophages using chromatin immunoprecipitation sequencing. Steroid Receptors: Methods and Protocols, 15-24.
- Park, P. J. (2009). ChIP–seq: advantages and challenges of a maturing technology. Nature reviews genetics, 10(10), 669-680.
Frontal image by <a href="https://www.freepik.es/foto-gratis/especialista-biotecnologia-laboratorio-realizando-experimentos_44133702.htm#query=ChIP%20seq%20DNA%20protein&position=0&from_view=search&track=ais">Freepik</a>
The moon, the plants, and the humans
How the story started
When it was proposed to do science outreach material as part of the EpiseedLink program, my first thought was to talk about curiosities about plants. Then, I asked my friends and family what they wanted to know about plants or which questions they had about plants. Then, my brother came up with a very interesting story.
In Brazil, we have a martial art called Capoeira, which mixes dance, music, and fighting. This martial art surged during the colonial period of the country and was developed by Africans brought to Brazil as slaves. Capoeira is closely related to the resistance of Africans and African descendants against slavery and the fight against racism. Nowadays, it is one of the symbols of Brazilian culture.
Capoeira uses different musical instruments and one of them is the berimbau. This instrument is mainly made of the wood of a tree called Biriba. When he was a Capoeira student, my bro ther heard his master saying that to make the instrument berimbau, you need to cut the tree in the evening, during the new moon or waning moon. The reason is that the probability of the wood rotting is lower if cut during this period.
ther heard his master saying that to make the instrument berimbau, you need to cut the tree in the evening, during the new moon or waning moon. The reason is that the probability of the wood rotting is lower if cut during this period.
This kind of knowledge is known as traditional knowledge, a result of observation, technologies, and practices developed by indigenous people, and passed down generation by generation. As a scientist, this made me very curious. How does the moon affect plant development? What do we know about this? Which other cultures made similar observations and have similar practices? Inspired by this story, I started my investigation to answer these questions.
The moon and its effect on animals
The most known effect of the moon on our planet is the tide. The gravity force of the moon pushes the water of oceans, increasing the water level. This happens twice a day, in a cycle known as the tidal cycle. The level of the seas increases more when the moon and the sun are aligned during the full and new moon. The gravitational force of these two celestial bodies sums up, generating what we call the spring tides. Also, the different moon phases  generate differences in the amount of light during the night, in a cycle of around 30 days.
generate differences in the amount of light during the night, in a cycle of around 30 days.
These differences in gravity and light due to the moon are detected by animals. For example, research indicates that the spawn of corals is dependent on moonlight intensity. Different species of nightjars follow the moon phases in their nesting period. The females of a crab species in Japan go down the mountain to lay their eggs during spring tides. These and other examples show the influence of the moon on animals' reproduction, physiology, and behavior.
The gravity effect on plants
What about the plants? One of the most known effects of gravitational force on plants being studied is the diameter of the trees and leaf movement. Under the natural variation of the light of day and night, the diameter of a tree increases more during the day, in a cycle of around 24h. However, if you put the tree in constant light, the diameter of the tree increases in a cycle of around 25h. This cycle is coincident with the variance in the local gravitational force. This local gravitational force is the result of the sum of the gravitational force of the moon and the sun, called the lunisolar gravitational force. It is supposed that when this force increases, it pulls the water in the tree, like the tides in the ocean.
As expected, the movement of the water in the trunk and the diameter of the truck seem to be correlated to the lunisolar gravitational force. It was also identified that the leaf movement of beans synchronized with the lunisolar gravitational force. If the plant can perceive the gravitational force, if there is a molecular mechanism associated with this, and how deep the effect of the lunisolar gravitational force is in the plant growth and development still needs to be investigated.
The light effect on plants
Even though the light of the moon is not enough to keep the plant alive, substituting the sun, this light seems to be detected by the plants. The moonlight is the result of the sunlight being reflected by the moon. However, the composition of these two lights is not the same. The moonlight is composed mainly of blue light in very low energy, while sunlight is composed of a long range of wavelengths at a high energy level. Then, it is expected that plants do not detect moonlight and sunlight equally.
The study of Breitler and collaborators investigates the detection of moonlight by plants from a molecular perspective. They discover that from the different proteins that detect light, known as photoreceptors, only one is expressed during full moon compared to new moon. They also detect many other genes that are affected by moonlight, having differences in their expression. What are the observable effects of the moonlight beyond the molecular effect must still be investigated. But since the moonlight affects animals, plants that are directly affected by animal predation, for example, may also use the moonlight as a cue.
The moon phases effect on plants
In Central America, more specifically Belize, Panama and Puerto Rico, indigenous people have the tradition of harvesting natural resources in the full moon. They perceive that the palm leaves collected during the full moon, for example, are more durable than the ones collected in other periods. These palm leaves are harvested for different reasons, one of the uses is to construct the rooftop of houses.
The study of Vogt and co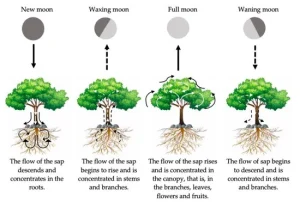 llaborators focuses on this tradition and tries to associate the full moon with the period when herbivores act more. Plants do not let themselves be consumed by herbivores passively. They defend themselves by producing chemical compounds. These chemical compounds can increase the durability of the plant and their survival in front of herbivores. What the scientists did was quantify chemical compounds associated with plant defense and durability in different moon phases. They identified that the concentration of some of the compounds quantified changed according to the moon phase. In the end, they suggest that plants produce more compounds against herbivores during the full moon. These compounds also help the durability of the leaves. The indigenous people, therefore, started to perceive that the durability increased during this period because the plant was defending itself from herbivores.
llaborators focuses on this tradition and tries to associate the full moon with the period when herbivores act more. Plants do not let themselves be consumed by herbivores passively. They defend themselves by producing chemical compounds. These chemical compounds can increase the durability of the plant and their survival in front of herbivores. What the scientists did was quantify chemical compounds associated with plant defense and durability in different moon phases. They identified that the concentration of some of the compounds quantified changed according to the moon phase. In the end, they suggest that plants produce more compounds against herbivores during the full moon. These compounds also help the durability of the leaves. The indigenous people, therefore, started to perceive that the durability increased during this period because the plant was defending itself from herbivores.
Conclusion
When to sow, when to harvest, when to cut the wood. Besides the period to cut the birimba to construct the musical instrument used in Capoeira or when to cut palm leaves to construct houses, a range of traditional human practices are correlated to the moon. Like the light and the temperature, the moon has a predictable cycle, inputting light and gravitational cues to the plants. Humans in different parts of the world observed this, giving rise to diverse traditional practices passed down from generation to generation in different human societies. Some of these practices were and are being studied by scientists. We are still in the initial stage of understanding how it all works, but gradually, the influence of the moon over plants will be unveiled.
Author: Maira Marins Dourado, PhD Student EpiSeedLink Marie Skłodowska-Curie Actions
Reviewed by: Ana Paula Avelino dos Santos
Curious? These are some of the references used in this article, that you may find interesting:
About traditional knowledge:
https://press.un.org/en/2019/hr5431.doc.htm
About Capoeira:
About human traditions, plants, and the moon:
http://www.soin-de-la-terre.org/wp-content/uploads/PlantsandtheMoon.pdf
About tides:
About animals and the moon:
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3712431/
About lunisolar gravity and plants:
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6215047/
About moonlight and plants:
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6961272/
About secondary metabolites and the moon:
https://www.jstor.org/stable/4315291
A recent review about the plants and the moon:
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC8866634/
Other references:
https://pubmed.ncbi.nlm.nih.gov/21002732/
http://www.soin-de-la-terre.org/wp-content/uploads/PlantsandtheMoon.pdf
https://link.springer.com/article/10.1007/s00709-010-0136-6
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6529214/#CIT0116
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6215041/
https://www.liebertpub.com/doi/epdf/10.1089/ast.2006.6.668
<a href="https://www.freepik.es/foto-gratis/hombres-entrenando-capoeira-playa_34305440.htm">Imagen de pikisuperstar</a> en Freepik












Brassica napus, known as oilseed rape, is a crucial crop prized for its oil-rich seeds and versatility. Yet, it faces challenges from pests and diseases in the field, impacting growth and quality. Exploring prevalent issues and effective management becomes pivotal for its cultivation's success.